Notice:
This post is older than 5 years – the content might be outdated.
In my previous blog post „how to manage machine learning models“ I explained the difficulties within the process of developing a good machine learning model and motivated using a tool to support data scientists with this challenge. This blog post will follow up by comparing three different tools developed to support reproducible machine learning model development:
- MLFlow developed by DataBricks (the company behind Apache Spark)
- DVC, an open-source project that gets its main support by the San Francisco-/ London based startup iterative.ai
- Sacred, an academical project developed by different researchers
Have a look at our 2022 update comparing frameworks for machine learning experiment tracking and see how MLflow, ClearML, neptune.ai and DAGsHub hold up!
First there will be one paragraph per framework that describes the project and shows some code examples. In the end of the article you will find a framework comparison and recommendations when to use which framework. As with my previous post the sklearn dataset on Boston-Housing prices will be used as basis. You can find a notebook to play with in this github repo. This notebook also includes instructions how to install the frameworks as well as some other functions we will use within the code examples below, but that won’t be discussed further, to place focus on the framework specific parts and omit boilerplate code.
DVC
DVC means „data (science) version control“ and aims to do for data science what git already does for software development: Making development processes traceable and reproducible. Therefore it strongly relies on git to track the source files and also uses it as an inspiration for its own CLI: To start a project, run „dvc init“, which will initialise some hidden files within a .dvc folder—just as git does. After initialisation you will need to write your program in such a way that everything will be stored within a file. DVC is agnostic to the programming language used, since it is controlled via terminal commands and does not have access to python variable values or similar.
DVC basically has three types of objects:
- dependencies: Everything your code relies on to run, such as other libraries, external scripts or the data source
- output files: Everything that will be created by your program and should be tracked, this can be preprocessing results as well as final predictions
- A metric file to store KPIs on the action taken place.
So the source code used in the example looks like this:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 |
def logDVC(model,param=dict(),metrics=dict(),features=None, tags=dict()): # Imports import json from sklearn.externals import joblib # Get some general information type = model.__module__.split(".")[0] # No option to set some tags to identify the experiment # Save Model if type=="sklearn": _ = joblib.dump(model,"tmp/mymodel") if type=="lgb": model.save_model("tmp/mymodel") # Log metrics with open('tmp/metrics.txt', 'w') as f: f.write(json.dumps(metrics)) # plot Feature importances if avaible plotFeatureImportances(model, features, type) # Create file about features if features is not None: with open("tmp/features.txt", "w+") as f: f.write(",".join(features)) if __name__ == "__main__": from sklearn.model_selection import train_test_split from sklearn.linear_model import Lasso # We need to import utils here, since it is an own script and the execution environment has no access to the jupyter execution environment from utils import * # Do a train_test_split data = getData() x_train, x_test, y_train, y_test = train_test_split(data.iloc[:,:-1], data.iloc[:,-1], test_size=10, random_state=42) # Define the details of our run params=dict(alpha=0.4) clf = Lasso(**params) clf.fit(x_train, y_train) predictions = clf.predict(x_test) metrics = eval_metrics(y_test, predictions) logDVC(clf,params,metrics,features=x_test.columns.values) |
We load the data, train an sklearn regression on it and store interesting information.
We will run the program like most other python programs but use the dvc run command in front to keep track of the results. Therefore we declare the three different types of objects we mentioned above:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
dvc run \ -d dvc_simple.py \ -f simple.dvc \ -o tmp/featureimportance.png \ -M tmp/metrics.txt \ -o tmp/features.txt \ -o tmp/mymodel python dvc_simple.py |
The f-flag specifies the name of the file dvc will use to track this command. Within this file it will store a hash value for alle mentioned files, so that dvc can track, if any of those have changed. With the command:
|
1 |
dvc repro simple.dvc |
you can reproduce the same experiment. DVC will check if any dependencies have changed and if so re-run the experiment. It will even detect if a dependent file was also specified as the output of another DVC run and if so, check those dependencies. This way you can build pipelines of different runs.
Using a command like
|
1 |
dvc metrics show |
you can check the results and compare them to an earlier run (but make sure you commited the state of the metrics file, since dvc will not do this out of the box for you)
Furthermore it is possible to version control even big dependent files such as input data using dvc. Therefore you should configure a remote storage such as a cloud bucket and then use the git-like syntax to add and push the relevant files:
|
1 2 3 4 5 |
dvc remote add myremote s3://bucket/path dvc add data.xml dvc push |
While DVC is great at reproducible experiments and sharing those with a team, it only provides little support to analyse the results. If you want to do ten runs using different settings, you will need to build some kind of comparison yourself. More complicated jobs with multiple dependencies and outputs make the cli quite verbose, but there are ideas to change this. Until those are implmenented, a good trick is to place all dependencies within a directory and then just link the whole folder. It is nice to be able to mix multiple languages but if you only use e.g. python anyway, it feels weird to write all information you want to store into files. Finally this design also hinders a tighter integration with notebooks, which makes dvc less usable for rapidly prototyping ideas.
MLFlow
When I first heard about Databricks creating MLFlow, I expected a heavy-weight tool with a tight spark integration, but I was completely wrong about that. MLFlow provides an API for python, R and Java as well as REST calls—of which I only tested the python version. It can be installed via pip and contains three sub-projects:
- Tracking: Function-calls for logging and a visualisation server (This is what we will focus on)
- Project: A definition of a project and dependencies to make them easily executable (we will only look at this briefly)
- Model: A standard for model storage and deployment (exploring this is left to you)
Let’s dive into the code. We’ll keep the main function similar and only use a new log-function:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 |
def logMlflow(model,data,output_folder="mlflow_out", param=dict(),metrics=dict(),features=None, tags=dict(),run_name=None): # Imports from sklearn.externals import joblib import mlflow import os if not os.path.exists(output_folder): os.makedirs(output_folder) # Get some general information type = model.__module__.split(".")[0] modelname = model.__class__.__name__ sha, remoteurl = getGitInfos() # Start actual logging mlflow.set_experiment(experiment_name="demo") if not run_name: run_name = modelname with mlflow.start_run(source_name=remoteurl,source_version=sha, run_name=run_name): # Log Parameters for k,v in param.items(): mlflow.log_param(k, v) # Track dependencies import pkg_resources with open("{}/dependencies.txt".format(output_folder), "w+") as f: for d in pkg_resources.working_set: f.write("{}={}\n".format(d.project_name,d.version)) mlflow.log_artifact("{}/dependencies.txt".format(output_folder)) # Track data data.to_csv("{}/data".format(output_folder)) mlflow.log_artifact("{}/data".format(output_folder)) if type=="sklearn": _ = joblib.dump(model,"{}/sklearn".format(output_folder)) mlflow.log_artifact("{}/sklearn".format(output_folder)) if type=="lgb": model.save_model("{}/lghtgbm.txt".format(output_folder)) mlflow.log_artifact("{}/lghtgbm.txt".format(output_folder)) # Log metrics for k,v in metrics.items(): mlflow.log_metric(k,v) # plot Feature importances if avaible featurePlot = plotFeatureImportances(model, features, type) if featurePlot: mlflow.log_artifact("{}.png".format(featurePlot)) # Set some tags to identify the experiment mlflow.set_tag("model",modelname) for tag, v in tags.items(): mlflow.set_tag(t,v) |
Using a with-statement combined with mlflow.start_run you can create a new run if there isn’t one yet and log the results of the experiment to it. Therefore three different log functionalities exist: Parameters for model configuration, metrics for evaluation and artifacts, for all files worth storage, input as well as output.
All this information is saved in a local folder named mlruns, which also is the information source for mlflows visualisation frontend. Running
|
1 |
mlflow ui |
will start the user interface that shows the different runs within an experiment as well as the parameters used and metrics created. You can also filter those based on parameters, metrics or tags. Furthermore it is possible to compare multiple runs in detail using a simple but effective scatter plot.
MLFlow does not offer a simple reproduce command such as DVC to re-run a predefined setting. How ever its „project“ component offers a way to increase standardisation. Therefore you can place a file called „MLProject“ into the root of you folder which serves as an entrypoint to your project. It looks like this:
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 |
name: tutorial conda_env: conda.yaml entry_points: main: parameters: alpha: float l1_ratio: {type: float, default: 0.1} command: "python train.py {alpha} {l1_ratio}" |
It references an environment file for dependencies and provides a main command with some modifiable parameters, in this case the alpha and the l1_ratio hyperparameter to control an ElasticNet model. Using the mlflow client you can then reproduce an experiment even without checking out its source code but directly accessing the github url:
|
1 |
mlflow run git@github.com:inovex/machine-learning-model-management.git -P alpha=0.4 |
In total, I found mlflow easy to understand due to its explicit logging functionality. Its integration does need a few extra lines of code, but its visualisation and comparison features are worth it, even in a quick prototyping phase.
Sacred
Sacred was developed in the academic environment and introduced at SciPy conference 2017. It only provides a python interface and uses annotations to define which parts and metrics to track. This way users don’t need to define every parameter that should be tracked manually and can thus minimise overhead. Sacred does not support capturing information developed within a notebook since the interactive nature of such notebooks makes code less reproducible, which is the primary goal of the framework. While you can test your Sacred code within a notebook, it does not really track the information but only checks the syntax. For this reason we again use the write_file magic of Jupyter to create a script from within the notebook and call it afterwards.
In the notebook you will find two versions of similar code to track experiments with Sacred. The first one is similar to the mlflow tracking code and uses dedicated logging functions. The second version (displayed below) is probably closer to the Sacred-way developers intended and tries to use annotations and special Sacred features.
|
1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 131 132 133 134 135 136 137 |
#!/usr/bin/env python from __future__ import division, print_function, unicode_literals from sacred import Experiment # Imports need to be done in the beginning of the file, since sacred won't recognize them, if they occur within a function from sklearn.externals import joblib ex = Experiment('Boston Housing Prices') from utils import * @ex.capture def capturestuff(_seed): print(_seed) def getData(): from sklearn.datasets import load_boston boston = load_boston() data = pd.DataFrame(boston.data,columns=boston.feature_names) data['target'] = pd.Series(boston.target) return data @ex.config def cfg(_log): alpha= 0.5 def logSacred(run,model,data,output_folder="sacred_out", param=dict(),metrics=dict(),features=None, tags=dict()): # Get some general information import os if not os.path.exists(output_folder): os.makedirs(output_folder) type = model.__module__.split(".")[0] modelname = model.__class__.__name__ # Config will be tracked automatically # Dependencies will also be tracked automatically # Track source code data.to_csv("{}/data".format(output_folder)) #ex.add_source_file("{}/data".format(output_folder)) ab = ex.open_resource("{}/data".format(output_folder)) # Create file about features if features is not None: with open("{}/features.txt".format(output_folder), "w+") as f: f.write(",".join(features)) ex.add_artifact("{}/features.txt".format(output_folder)) # plot Feature importances if avaible if plotFeatureImportances(model, features, type): ex.add_artifact("{}/featureimportance.png".format(output_folder)) # Track Model binary if type=="sklearn": _ = joblib.dump(model,"{}/sklearn".format(output_folder)) ex.add_artifact("{}/sklearn".format(output_folder)) if type=="lgb": model.save_model("{}/lghtgbm.txt".format(output_folder)) ex.add_artifact("{}/lghtgbm.txt".format(output_folder)) # Log metrics for k,v in metrics.items(): ex.log_scalar(k,v) # Set some tags to identify the experiment for tag, v in tags.items(): ex.add.set_tag(t,v) @ex.automain def run(_run, alpha): # Setup from sklearn.model_selection import train_test_split from sklearn.linear_model import Lasso from sklearn.metrics import mean_absolute_error # Do a train_test_split on my Data data = getData() x_train, x_test, y_train, y_test = train_test_split(data.iloc[:,:-1], data.iloc[:,-1], test_size=10, random_state=42) # Define my params params=dict(alpha=alpha) clf = Lasso(**params) clf.fit(x_train, y_train) predictions = clf.predict(x_test) metrics = eval_metrics(y_test, predictions) logSacred(_run,clf,data,param=params,metrics=metrics,features=x_test.columns.values) |
Maybe it is just because I use Sacred for the first time and already wrote the code for the two other frameworks, but using the Sacred annotations did not feel very natural. And while the Sacred magic of auto-collecting important information seems compelling at first, it is far from perfect and I found myself frequently controlling whether my dependencies and files got tracked properly. For example Sacred will track dependencies if you import them at the top of your script, but not if you do this within a function. It also provides some commands such as open_resource to read files and track them at the same time, but you will need to remember those special commands and if your data is not stored in a file but received from an API for example you will need to log the dependency explicitly nevertheless. To me, the Sacred way of annotating the code was more of a burden than a relieve compared to explicit logging.
That said, Sacred probably is a good candidate if you want to create your own framework for your organisation, since sacred uses a modular design approach. Its API provides many functions and hooks to create custom wrappers of your own. While the number of options will overwhelm normal developers that just want to keep track of some libraries and dependencies, it will give you the power to shape the tool according to your needs if you want to do so. Not only can its functionality be extended in multiple ways, Sacred also supports different storage backends like file storage, MongoDB, TinyDB, SQL and others. The examples in the notebook use MongoDB since this is the only one supported by the existing visualisation tools, which are external components as well.
While none of the three listed visualisation tools offers as much functionality as mlflow, the different backends should make it easy to create your own visualisation as well. The screenshots below show omniboard which allows you to create new columns for metrics logged and supports ordering, but lacks a comparison of different runs in detail.
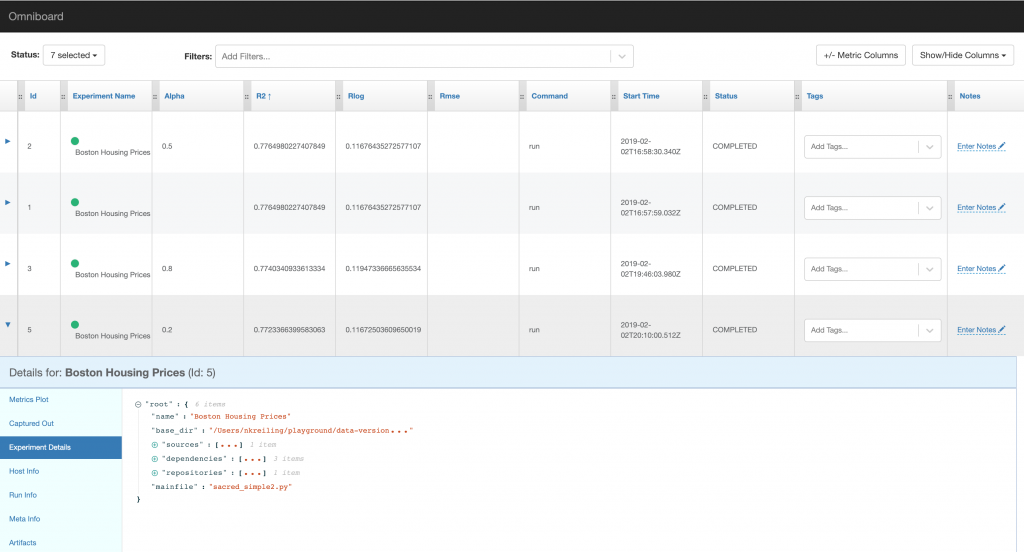
Conclusion
The three different frameworks leverage different purposes. While DVC targets universal applicability and prioritises reproducibility over ease-of-use, mlflow is a nice and convenient tool to track your parameters and compare model performance while prototyping. Sacred’s use case is somewhat limited, it only provides a python interface, does not support usage within notebooks and does not offer a simple command to reproduce an experiment. Nevertheless it shines with its open standards and extensibility.
| DVC | mlFlow | Sacred | |
| Language Support | Universal using Shell | Python, R, Java, REST | Python |
| Control | DAG using Scripts | Python Package | Annotations |
| Storage Backend | File storage | File storage | MongoDB + others |
| Github Stars | 2400 | 3400 | 1700 |
| License | Apache 2 | Apache 2 | MIT |
(last updated 4.4.2019)
If you are looking for a tool that gives you an easy way to log important attributes and helps you organise your models while prototyping, go for mlflow.
If you work in a team and want to make your experiments absolutely reproducible even if it takes some extra steps: check out DVC.
Delve into Sacred if you want to build your own tool and profit from previous work, or if the others don’t offer you enough flexibility.
| Features | DVC | mlFlow | Sacred |
| Notebook support | Yes | Yes | Only Dry-Run |
| model storage | Yes | As Artefacts | Only Artefacts |
| Input data storage | Yes | Only Artefacts | Yes |
| Visualisation | No | Yes | Yes (External) |
| Parameter Tracking | Yes | Yes | Yes |
| Performance Metrics | Yes | Yes | Yes |
| Execution statistics | Little | Little | Yes |
| Source Code Tracking | Via Git | Manually (Git Hashes) | Yes |
| Dependencies | Manually (Env-File) | Manually (Env-File) | Yes |
Read on
Find our complete Data Science portfolio on our website or have a look at our job openings and consider joining us as a data scientist!



Hey, one of DVC authors is here. Great blogpost! ML folks often ask what is the difference between DVC.org and MLflow.org. Thank you!
A few details:
1. the screenshot is very small – I don’t see it even with my glasses 🙂
2. „DAG von Skripten“ German language?
3. „DVC, a software product of the London based startup iterative.ai“ Our HQ and founders are in San Francisco actually. But the team is distributed. I won’t be surprised if a middle point of all the team members is somewhere around London as an average. So, you can keep London :))) And DVC was started as a pet project by enthusiasts and it was turned into a startup by these enthusiasts a bit latter.
4. Number of GutHub stars is outdated. DVC has 2300 today, mlflow – 3000+.
Great post. For those looking for aproduction ready product take a look at Comet.ml. We’ve been using it for the past six months and it’s amazing. Lots of similarities with Mlflow but much more mature.